The Segmentation tool is a method for simple mask creation similar to the segmentation in the 3D tool. It opens the following dialog window.
The following segmentation methods are available.
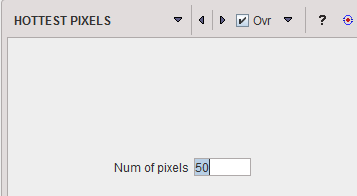
HOTTEST |
The HOTTEST PIXELS segmentation allows obtaining the (often disconnected) pixels with the highest values. The number of included pixels can be specified in the Num of pixels field.
|
REGION |
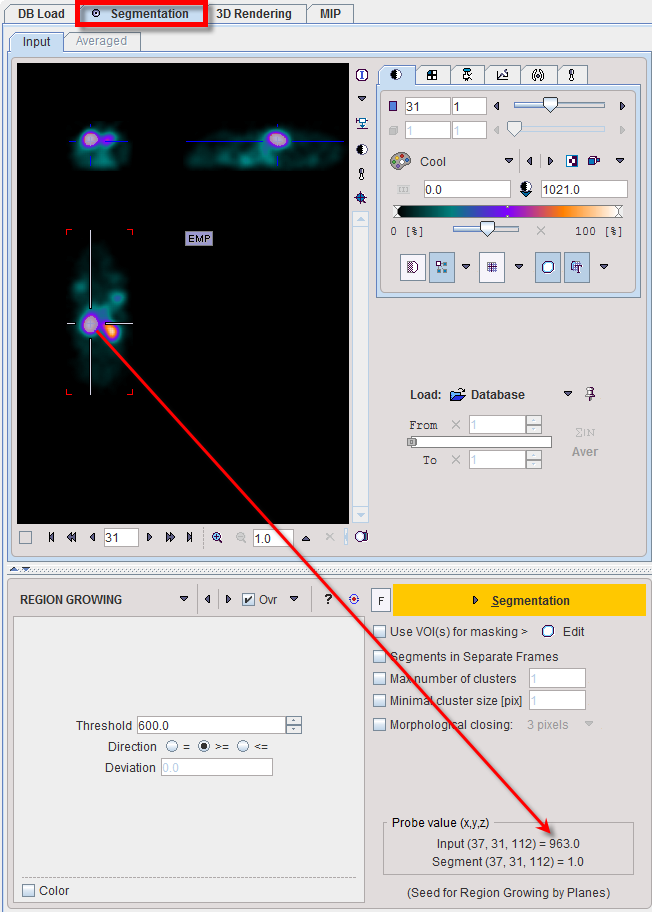
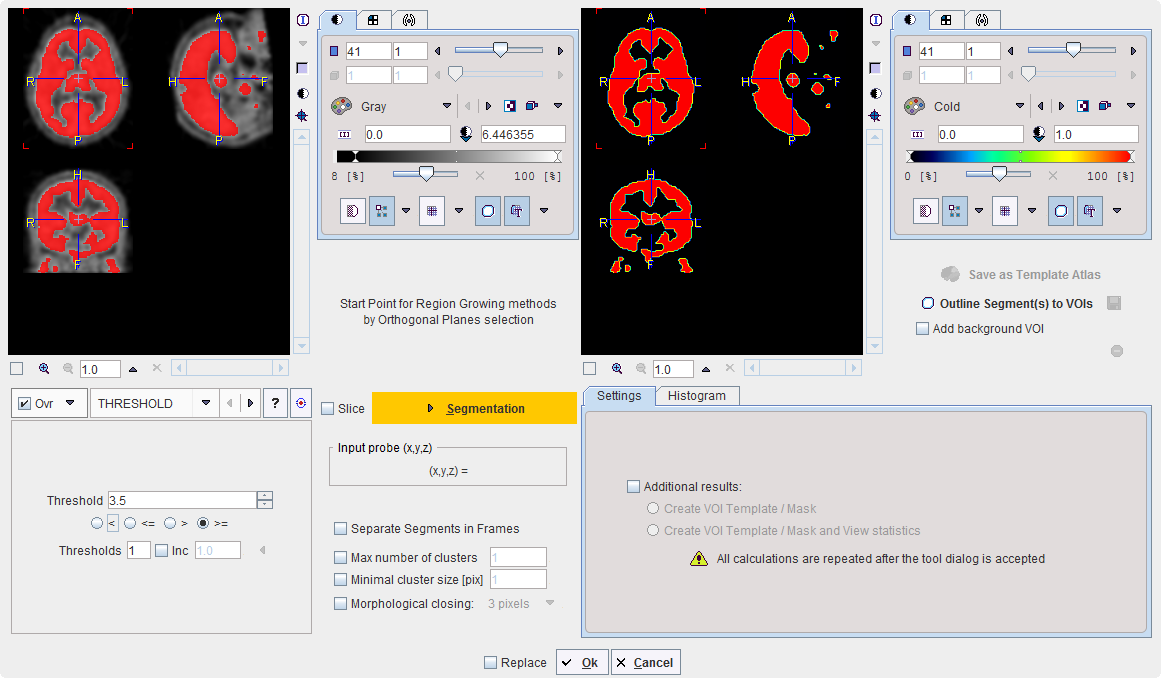
REGION GROWING is a method by which the user defines a starting point (seed) within the object of interest, and the algorithm tries to find all connected pixels which fulfill a certain criterion. It is required to triangulate the seed point with the orthogonal plane layout as illustrated below.
The inclusion criterion is formed by the Threshold value and the Direction specification: ▪= include all connected pixels with a value Threshold ± Deviation; ▪>= include all connected pixels above the defined Threshold value; ▪<= include all connected pixels below the defined Threshold value. Note the Probe value which displays the image value at the position of the cursor, which is useful for finding appropriate Threshold setting. |
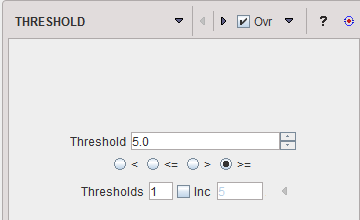
THRESHOLD |
The THRESHOLD segmentation is conceptually simple. All pixels with value above or below the threshold are included in the segment, depending on the condition settings.
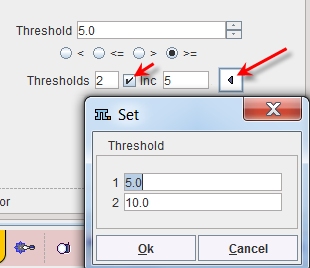
Sometimes it is helpful to segment at several threshold levels at once. This can easily be realized by setting the number in the Thresholds field accordingly, and entering the threshold values in the appearing Set dialog window.
Multiple threshold values can be incremented with a constant value enabling the Inc box and specifying the step in the dedicated text box. |
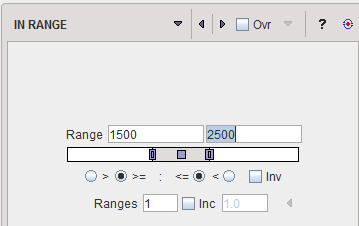
IN RANGE |
The IN RANGE segmentation is similar to the THRESHOLD method, except that two limits are defined. The boundary values are included or excluded, depending on the radio button settings. The complementary criterion is obtained by checking the Invert box.
Similar to the THRESHOLD segmentation, multiple Ranges can be defined and segmented at once. |
Otsu Threshold (ITK) |
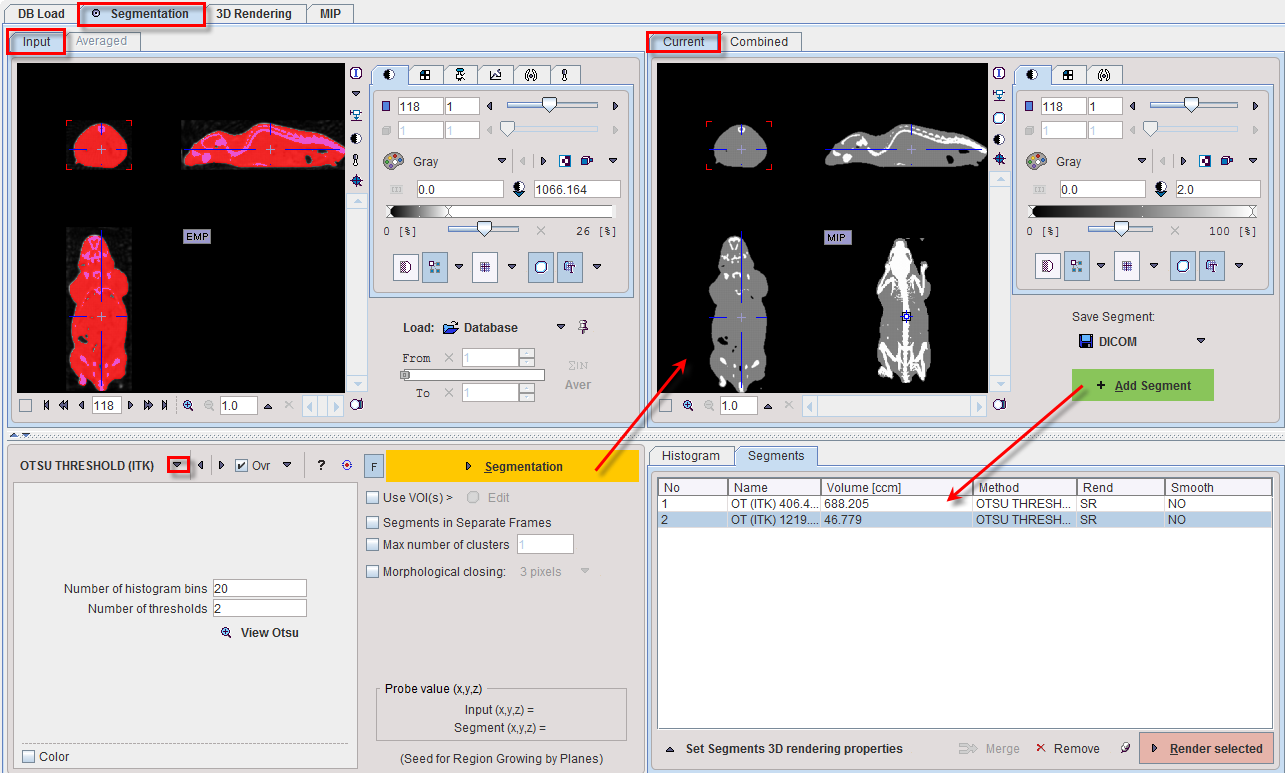
The OTSU THRESHOLD method is based on the analysis of the pixel histogram and is described by The ITK Software Guide as follows: "Another criterion for classifying pixels is to minimize the error of misclassification. The goal is to find a threshold that classifies the image into two clusters such that we minimize the area under the histogram for one cluster that lies on the other cluster’s side of the threshold. This is equivalent to minimizing the within class variance or equivalently maximizing the between class variance." The Number of histogram bins defines the resolution of the histogram shape, whereas the Number of thresholds defines the number of distinct clusters. With the mouse CT, Otsu with the settings below finds appropriate thresholds for the skeleton object and the soft tissue, as illustrated below. Note that the threshold values can be inspected only after the actual segmentation by the View Otsu button.
|
Neighborhood Connected (ITK) |
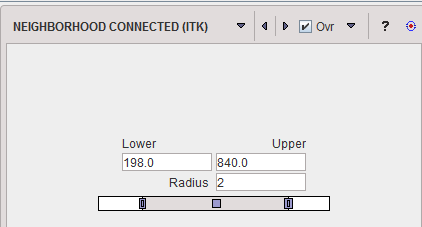
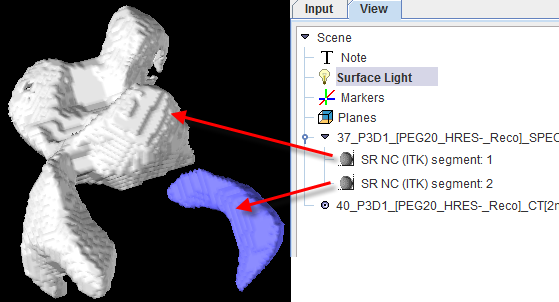
NEIGHBORHOOD CONNECTED is a region growing method which applies two grouping criteria: 1.An included pixel must have a value in the range between the Lower and Upper threshold. 2.All pixels in its neighborhood within the specified pixel Radius must also be within the value range. By the combination of these criteria small structures are less likely to be accepted in the region.
A single seed is provided by the triangulation point of the orthogonal planes. Multiple seed points can be specified by the use of markers.
The number of structures found depends on the number of seeds.
|
Confidence Connected (ITK) |
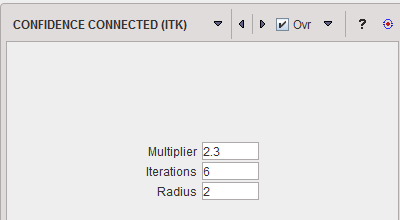
From the The ITK Software Guide: "The criterion used by the Confidence Connected method is based on simple statistics of the current region. First, the algorithm computes the mean and standard deviation of intensity values for all the pixels currently included in the region. A user-provided factor (Multiplier)is used to multiply the standard deviation and define a range around the mean. Neighbor pixels whose intensity values fall inside the range are accepted and included in the region. When no more neighbor pixels are found that satisfy the criterion, the algorithm is considered to have finished its first iteration. At that point, the mean and standard deviation of the intensity levels are recomputed using all the pixels currently included in the region. This mean and standard deviation defines a new intensity range that is used to visit current region neighbors and evaluate whether their intensity falls inside the range. This iterative process is repeated until no more pixels are added or the maximum number of iterations is reached. The number of Iterations is specified based on the homogeneity of the intensities of the anatomical structure to be segmented. Highly homogeneous regions may only require a couple of iterations. Regions with ramp effects, like MRI images with inhomogeneous fields, may require more iterations. In practice, it seems to be more important to carefully select the multiplier factor than the number of iterations. However, keep in mind that there is no reason to assume that this algorithm should converge to a stable region. It is possible that by letting the algorithm run for more iterations the region will end up engulfing the entire image." The initialization of the algorithm requires the user to provide a seed point. It is convenient to select this point to be placed in a typical region of the anatomical structure to be segmented. A small initial neighborhood Radius around the seed point will be used to compute the initial mean and standard deviation for the inclusion criterion.
|
Connected |
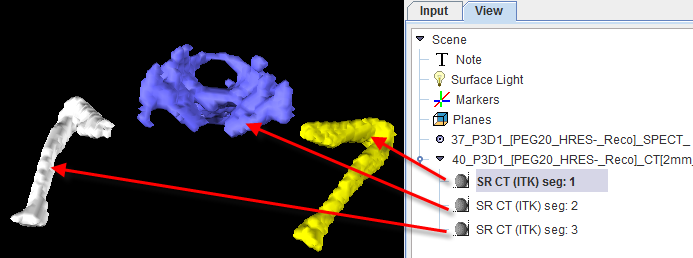
Connected Threshold is a region growing method with the criterion that (similar to the IN RANGE method) the included pixels must have values between a Lower and Upper threshold.
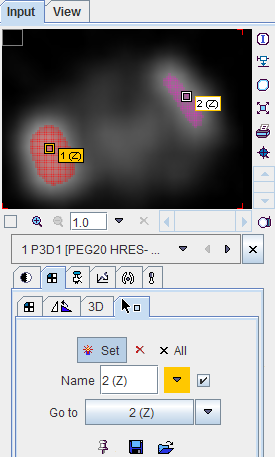
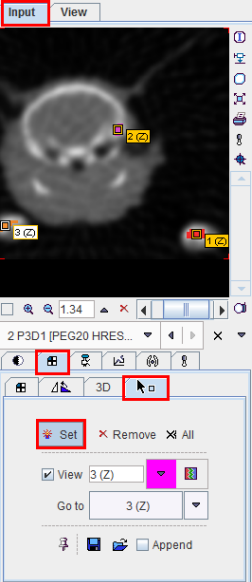
As usual with region growing methods, the user has to specify a seed point by clicking into the image. The ITK region growing methods are particular in that multiple seed points can be specified at once by the use of markers as illustrated below.
Once the markers tab has been activated and the Set button enabled, a marker is created as a seed point for each clicking into the image. When the segmentation is started, the region growing with the specified value range is performed from each marker.
|
SCATTER in VOI |
This particular method for localizing pixels from 2D scatter plots is described in a separate section. |
After specification of the segmentation method and its parameters, the segmentation can be started with the Segmentation button. The result is shown in the right image display port. Note that if the study is dynamic, an additional box All Frames appears. If it is checked, segmentation will be performed for all frames separately, otherwise with the frame which is currently shown.
The segmentation result can be used in different ways:
▪Save as Template Atlas directly saves the segmented series in an atlas format in the directory Pmod4.1/resources/templates/voitemplates. Subsequently, it can be used for statistics using the Atlas tab of the Templates pane in the VOI definition function.
▪The Outline button allows creating VOIs for each generated segment. Optionally, the background can be added as a VOI by enabling the Add background VOI box. The outlined VOIs can be saved as files and later used for statistics by the Save icon next to the Outline button.
With the Ok button the segmentation is performed again and the resulting segment returned as an image series which is binary for a simple segmentation and an object map in the case of a clustering method. The following Additional results can be returned.
1.Create VOI Template/Mask: With this setting, the segments are also returned as a VOI template which can be activated by the Mask tab of the VOI Template function.
2.Create VOI Template/Mask and View Statistics: As 1), but the statistics on the current image are additionally calculated.