The MR page allows loading the T1-weighted brain MR image of the same subject using the Load T1 MR button. The MR image has to appear in the HFS orientation as illustrated below. If there are two matched MR images with different contrasts (e.g. T1 and T2) both can be loaded and used for a multi-spectral segmentation using the 6 Probability Maps (SPM12) method.
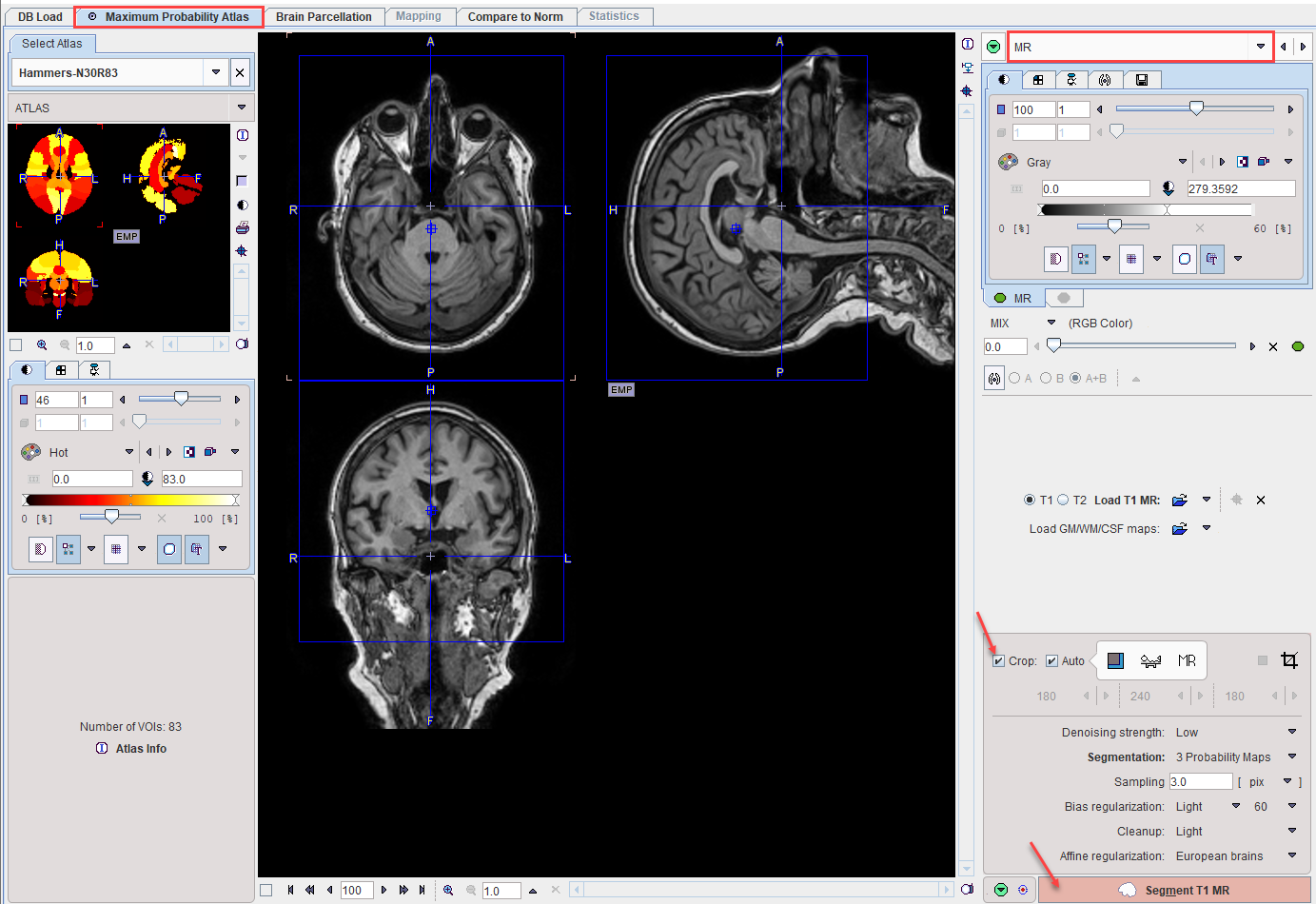
MR Image Cropping
The MR image will be the basis for determining the stereotactic normalization. Experience has shown that problems may occur if the MR field-of-view is much larger than the template as occurs for instance with sagittal MR acquisitions. Therefore, please use the Crop facility as described before for reducing the MR data set to the relevant portion with skull and brain, but without the neck.
MR Image Segmentation
The MR image will be segmented into gray matter (GM), white matter (WM) and cerebrospinal fluid (CSF). The algorithm uses the following parameters:
Denoising strength |
Denoising of the MR image may improve the segmentation of gray matter, white matter and CSF. If a Denoising strength other than None is selected, a non-local means denoising algorithm is applied which preserves structure boundaries unless the strength is too high. |
Segmentation |
Two SPM-type segmentation variants are supported, the 3 Probability Maps (SPM8) and the 6 Probability Maps (SPM12) variant. Note that the normalization transform which is obtained as part of the segmentation can be applied for the spatial normalization of the subject brain anatomy in later stages. |
Sampling |
Density of pixels considered in the calculation. It can be specified in pixel or mm units. |
Bias regularization |
Serves for compensating modulations of the image intensity across the field-of-view. Depending on the degree of the modulation, a corresponding setting can be selected from the list. The parameter to the right indicates the FWHM [mm] to be applied. The larger the FWHM, the smoother the variation is assumed. |
Cleanup |
Procedure for rectifying the segmentation along the boundaries. |
Affine regularization |
Two different initializations of the affine registration are supported, European brains and East Asian brains, as well as No regularization. The setting should correspond to the nature of the subject under study. |
It is recommended to use the default settings and only experiment with other parameter values if the segmentation fails. The default settings can be recovered by the ![]() button.
button.
The actual segmentation is started with the Segment T1 MR action button. Note that the denoising and segmentation process may take several minutes.