The Kinetic Modeling tool in the pipeline has the following interface. Please refer to PMOD Kinetic Modeling Users Guide for details regarding the modeling configuration.
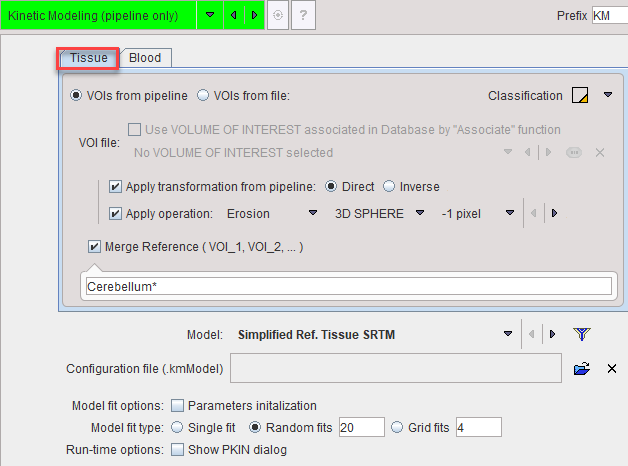
Volumes of Interest for Tissue TACs
The kinetic tool generates the average tissue TACs from VOIs which are applied to the current pipeline data. They are specified on the Tissue tab and can be provided in three ways:
▪VOIs from pipeline: The last VOIs produced in the pipeline are applied.
▪VOIs from file: One option is to use VOIs which have been produced previously and associated to the current pipeline images.
![]()
▪VOIs from file: Another option is to use a fixed VOI set.
![]()
This is particularly useful for data which have been transformed to a template space, such as the MNI space.
The selected VOIs can optionally be transformed before application to the images:
▪Apply transformation from pipeline: The idea here is that the VOIs have been derived using an image different from the current images, and that the pipeline has recorded the appropriate transformation for bringing the VOIs into the space of the current image. Such an application is described in the example below. The Direct or the Inverse transformation can be applied, as appropriate.
▪Apply Operation: The operations offered for VOI modification are the Erosion or Dilation by a number of pixels. Erosion allows shrinking VOIs from the outside. This may be useful for reducing the partial-volume effect. Dilation grows the VOIs by adding additional layers.
As a convenience for applying reference tissue models a sub-set of VOIs can be merged with the Merge Reference VOI option. The specification is by a comma-separated list of exact VOI names like Cerebellum_l, Cerebellum_r, or using the "*" wildcard character such as Cerebellum* to enclose all VOI names starting with "Cerebellum".
Blood Activity Information
If blood-based kinetic models are applied in pipeline processing, blood information must be available for each data set processed. Unless the information is common among data sets, the blood information must have been associated to the PET data beforehand. The Blood tab needs to be selected for the specification as illustrated below. In this example the Whole blood activity and the Plasma activity are referenced by association, whereas a constant Parent Fraction is applied which assumes that the metabolite buildup is common among the population processed.
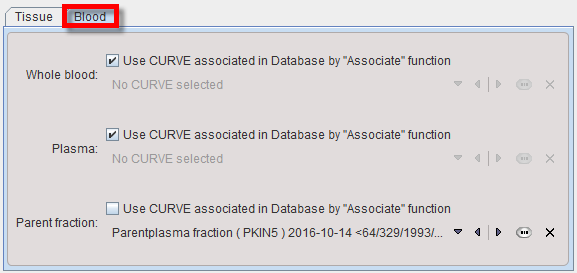
Not that at this time the plasma fraction variant for specification of the plasma activity is not supported in pipeline processing.
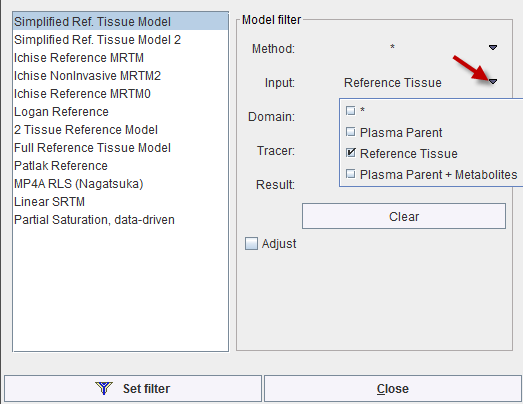
Model Selection
The tissue model to be applied in fitting can be selected from the Model list.
![]()
Because of the large model number the list is initially filtered to only show reference tissue models. Please use the filter button indicated above for showing the filter setting window. There, filtering can be switched off with the Clear button, or set to different criteria. Set filter closes the dialog window and updates the Model list.
Fitting Options
Each tissue model has its default regarding initial parameters and fitting flags. These defaults can be changed in the kinetic tool (PKIN). Alternatively, an explicit model configuration file can be prepared beforehand in PKIN and selected as the Configuration file.
![]()
The Model fit options allow applying some of the fitting options available in interactive modeling. Briefly:
▪Parameters initialization: This option is only effective for compartment models. If enabled, the initial parameters are calculated using a linear least squares method.
▪Random fits: If the option is enabled, the regional fits are repeated the specified number of times with randomly varied initial parameters. In the end, the fit with minimal Chi square will be returned as the result.